Transcription of Chapter 14. Enzyme Kinetics
1 Chapter 14. Enzyme Kinetics Chemical Kinetics Elementary reactions A P (Overall stoichiometry). I1 I2 (Intermediates). Rate equations aA + bB + +zZ P. Rate = k[A]a[B]b [Z]z k: rate constant The order of the reaction (a+b+ +z): Molecularity of the reaction Unimolecular (first order) reactions: A P. Bimolecular (second order) reactions: 2A P or A + B P. Termolecular (third order) reactions Rates of reactions A P (First-order reaction). d[P] d[A]. v= =- = k[A]. dt dt 2A P (Second-order reaction). d[P] d[A]. v= =- = k[A]2. dt dt A + B P (Second-order reaction). d[P] d[A] d[B]. v= =- =- = k[A][B]. dt dt dt Rate constant for the first-order reaction d [A]. v= = k[A]. dt d [A]. = kdt [A]. [ A] d [A] t [A]0 [A] = k 0 dt ln[A] ln[A]0 = kt ln[A] = ln[A]0 kt [A] = [A]0 e kt (t). The reactant concentration decreases exponentially with time Half-life is constant for a first-order reaction ln[A] ln[A]0 = kt [A].
2 Ln = kt [A]0. 1. ln = kt 1 / 2. 2. ln 2 t1 / 2 = =. k k For the first-order reaction, half-life is independent of the initial reactant concentration Second-order reaction with one reactant 2A P. d [A]. v= = k [ A ]2. dt d [A] 1 Slope= k = kdt [A] 2 [A]. [ A] d [A] t [A]0 [A]2 = k 0 dt 1. 1 1. = + kt [A]0. [A] [A]0. Time (t). 2 1 1. kt 1 / 2 = =. [A]0 [A]0 [A]0. 1. t1 / 2 = Half-life is dependent on the initial reactant concentration k[A]0. Pseudo first-order reactions v = k[A][B]. When [B] >> [A], v = k [A]. where k = k[B]0. B is water ( ): k'= Arrhenius equation k = Ae-Ea/RT. = Activation energy (Ea). Multistep reactions have rate-determining steps k1 > k2. k1 < k2. k1 k2. Rate-determining step: the slow step (= the higher activation energy). Catalysts reduce the activation energy Ea (the reduction in Ea by the catalyst). Rate enhancement = kcat/kuncat = e Ea/RT.
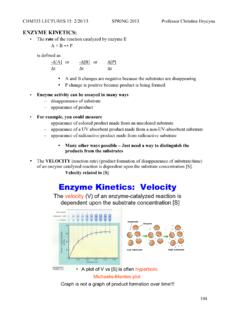
3 When Ea = kJ/mol, the reaction rate increases 10 fold When Ea = 34 kJ/mol, the reaction rate increases 106 fold Michaelis-Menten equation Vmax [S]. v0 =. K M + [S]. v0 = the initial rate Vmax = the maximum rate KM = the Michaelis constant [S] = the substrate concentration Steady state approximation k1 k2. E+S ES P+E. k-1. Derivatization of Michaelis-Menten equation k1 k2. E+S ES P+E. k-1. d [ P]. v= = k 2[ES]. dt d [ES]. = k 1[E][S] k 1[ES] k 2[ES] = 0 (Steady state approximation). dt [E]T = [E] + [ES]. k 1([E]T [ES])[S] = (k 1 + k 2)[ES]. (k 1 + k 2 + k 1[S])[ES] = k 1[E]T[S]. k 1 + k2. ( + [S])[ES] = [E]T[S]. k1. [E]T[S]. [ES] =. k 1 + k2. + [S]. k1. [E]T[S] k 1 + k2. [ES] = where KM =. KM + [S] k1. k 2[E]T[S]. The initial velocity v 0 = k 2[ES] =. KM + [S]. The maximum velocity (Vmax) occurs at high substrate concentration when the Enzyme is entirely in the [ES] form : Vmax = k 2[E]T.
4 Vmax [S]. v0 =. K M + [S]. Michaelis constant KM. KM = [S] at which v0 = Vmax/2. If an Enzyme has a small value of KM, it achieves maximal catalytic efficiency at low substrate concentrations Measure of the Enzyme 's binding affinity for the substrate (The lower KM, the higher affinity). Lineweaver-Burke plot Vmax [S]. v0 =. K M + [S]. 1 K M + [S]. =. v 0 Vmax [S]. 1 KM 1 1. = +. v 0 Vmax [S] Vmax kcat/KM is a measure of catalytic efficiency Catalytic constant or turnover number of an Enzyme : Vmax kcat =. [E]T. When KM >> [S], very little ES is formed and consequently [E] [E]T. Vmax[S] Vmax[S] kcat[E]T[S] kcat . v0 = = [E][S]. KM + [S] KM KM KM . Catalytic perfection kcat k 2 k 1k 2. = =. KM K M k 1 + k 2. The ratio is maximal when k 2 >> k 1, kcat = k1. KM. (Diffusion-controlled limit: 108 to 109 M-1s-1). Inhibitors Substances that reduce an Enzyme 's activity Study of enzymatic mechanism Therapeutic agents Reversible or irreversible inhibitors O CO2- O CO2- O N CO2- H NH2 N CO2- N H.
5 HN N N. N N. H. H2N N N CH3. H2N N N. H. Dihydrofolate Methotrexate (Dihydrofolate reductase substrate) (Dihydrofolate reductase inhibitor, anticancer drug ). Modes of the reversible inhibition Competitive inhibitors Binds to the substrate binding site Uncompetitive inhibitors Binds to Enzyme - substrate complex Non-competitive inhibitors Binds to a site different from the substrate binding site Mixed inhibitors Binds to the substrate- binding site and the Enzyme -substrate Competitive inhibition [E][I]. KI =. [EI]. d [ES]. = k 1[E][S] k 1[ES] k 2[ES] = 0 (Steady state approximation). dt k 1 + k 2 [ES] KM [ES]. [E] = =. k 1 [S] [S]. [E][I] KM [ES][I]. [EI] = =. KI [S]KI. [E]T = [E] + [EI] + [ES]. KM [ES] KM [ES][I] KM [I] . [E]T = + + [ES] = [ES] 1 + + 1 . [S] [S]KI [S] KI . [E]T[S]. [ES] =. [I] . KM 1 + + [S]. KI . k 2[E]T[S]. v 0 = k 2[ES] =. [I] . KM 1 + + [S].
6 KI . Since Vmax = k 2[E]T, Vmax [S] [I]. v0 = where = 1 +. K M + [S] KI. Competitive inhibitors affect KM. Vmax [S]. v0 =. K M + [S]. [S] ; v0 Vmax regardless of . Determination of KI. of the competitive inhibitor 1 KM 1 1. = +. v0 V max [S] V max Uncompetitive inhibition [ES][I]. KI ' =. [ESI]. KM [ES]. [E] =. [S]. [ES][I]. [ESI] =. KI '. [E]T = [E] + [ES] + [ESI]. KM [ES] [ES][I] KM [I] . [E]T = + [ES] + = [ES] + 1 + . [S] KI ' [S] KI ' . [E]T[S]. [ES] =. [I] . KM + 1 + [S]. KI ' . k 2[E]T[S]. v 0 = k 2[ES] =. [I] . KM + 1 + [S]. KI ' . Since Vmax = k 2[E]T, Vmax [S]. Vmax [S] ' [I]. v0 = = where ' = 1 +. K M + '[S] K M + [S] KI '. '. Uncompetitive inhibitors decrease both Vmax and KM. Vmax [S]. v0 = '. KM. + [S]. '. Vmax [S] ; v0 . '. V (Effects of an uncompetitive inhibitor K M >> [S]; v0 max [S]. KM become negligible). Vmax no inhibitor ( '=1). Increasing [I].
7 VmaxI. v0 Vmax/2. [I]. VmaxI/2 ' = 1+. KI '. KMI KM [S]. Determination of KI'. of the uncompetitive inhibitor 1 KM 1 '. = +. v0 V max [S] V max Mixed inhibition [E][I] [ES][I]. KI = KI ' =. [EI] [ESI]. [E][I]. [EI] =. KI. [ES][I]. [ESI] =. KI '. [E]T = [E] + [EI] + [ES] + [ESI]. [I] [I] . [E]T = [E] 1 + + [ES] 1 + . KI KI ' . [E]T = [E] + [ES] '. KM [ES] KM . [E]T = + [ES] ' = [ES] + ' . [S] [S] . [E]T[S]. [ES] =. KM + '[S]. k 2[E]T[S]. v 0 = k 2[ES] =. KM + '[S]. Since Vmax = k 2[E]T, Vmax [S] [I] [I]. v0 = where = 1 + and ' = 1 +. K M + '[S] KI KI '. Lineweaver-Burk plot of a mixed inhibition 1 KM 1 '. = +. v0 V max [S] V max Noncompetitive inhibition A special mixed inhibition when KI = KI'. Vmax [S] [I] [I]. v0 = where = 1 + and ' = 1 +. K M + '[S] KI KI '. When KI = KI' , = '. Vmax [S]. Vmax [S]. v0 = = . ( K M + [S]) K M + [S]. Noncompetitive inhibitors affect not KM but Vmax Vmax [S].
8 [ I]. v0 = where = 1 +. K M + [S] KI. Vmax [S] ; v0 .. Vmax no inhibitor ( =1). Vmax I Increasing [I]. v0 Vmax/2. VmaxI/2 [I]. = 1+. KI. KM [S]. Determination of KI. of the noncompetitive inhibitor 1 KM 1 . = +. v0 V max [S] V max 1/v0. =2. Increasing = [I]. = 1. (no inhibitor). Slope= KM/Vmax [ I]. /Vmax = 1+. KI. 1 0 1/[S].. KM. Effects of inhibitors on Vmax and KM. of the Michaelis-Menten equation app Vmax [S]. v0 = app K M + [S]. Effects of pH. '. Vmax [S]. Binding of substrate to Enzyme v0 = '. K M + [S]. Catalytic activity of Enzyme '. Vmax = Vmax / f 2 and K M' = K M ( f 1 / f 2). Ionization of substrate where Variation of protein structure [H + ] KE2. f1= +1+ +. KE1 [H ]. (only at extreme pHs). [H + ] KES2. f2= +1+ +. KES1 [H ]. E- ES- KE2 H+ KES2 H+. k1 k2. EH + S ESH P + EH. k-1. KE1 H+ KES1 H+. EH2+ ESH2+. Approximate identity of catalytic amino acid residues pKa ~4 Catalytic Asp or Glu residue pKa ~6 Catalytic His residue pKa ~10 Catalytic Lys residue Caution should be taken because pKa of amino acid residues are environmentally sensitive Bisubstrate reactions 60% of biochemical reactions involve two substrates and two products Transfer reactions and oxidation-reduction reactions Cleland's nomenclature system for the enzymatic reactions Substrates: A, B, C, D in the order that they add to the Enzyme Products: P, Q, R, S in the order that they leave the Enzyme Inhibitors: I, J, K, L.
9 Stable Enzyme complexes: E, F, G, H with E being the free Enzyme Numbers of reactants and products: Uni (one), Bi (two), Ter (three), and Quad (four). Bi Bi reaction: a reaction that requires two substrates and yields two products Types of Bi Bi Reactions Sequential reactions (single displacement reactions): all substrates bind before chemical event Ordered mechanism Random mechanism Ping pong reactions (double displacement reactions): chemistry occurs prior to binding of all substrates Differentiating bisubstrate mechanisms Measure rates Change concentration of substrates and products Lineweaver-Burk plot Intercept (1/Vmax): the velocity at saturated substrate concentration It changes when the substrate A. binds to a different Enzyme form with the substrate B. Slope (KM/Vmax): the rate at low substrate concentration It changes when both A and B. reversibly bind to an Enzyme form Ping Pong Bi Bi Mechanism Intercept changes because A and B bind to the different Enzyme forms E and F, respectively Slope remains same because the binding of A and B is irreversible due to the release of the product (P).
10 Sequential Bi Bi Mechanism or Ordered Random Intercept changes because A and B binds to the different Enzyme forms (E or EB) and (EA or E), respectively Slope changes because the binding of A and B is reversible Differentiating Bi Bi mechanisms by product inhibition Competitive inhibition Substrate and inhibitor competitively bind to the same site of the Enzyme A vs Q: Competitive B vs P: Competitive Ping Pong A vs P: Noncompetitive B vs Q: Noncompetitive A vs Q: Competitive B vs P: Noncompetitive Ordered A vs P: Noncompetitive sequential B vs Q: Noncompetitive Under assumption of dead-end complex formation (A is similar with Q and B is similar with P). Random A vs Q: Competitive sequential B vs P: Competitive A vs P: Noncompetitive B vs Q: Noncompetitive Dead-end complexes A B. E. Q P. Q B Dead-end complex (no chemistry). A P no chemistry competitive ATP + Creatine ADP + Creatine-phosphate competitive Differentiating Bi Bi mechanisms by isotope exchange Ping Pong Mechanism A* P isotope exchange is possible without B.