Transcription of Chapter 13: Spectroscopy - Vanderbilt University
1 11 Chapter 13: SpectroscopyMethods of structure determination Nuclear Magnetic Resonances (NMR) Spectroscopy (Sections ) Infrared (IR) Spectroscopy (Sections ) ultraviolet - visible (UV-Vis) Spectroscopy (Section ) Mass (MS) spectrometry (not really Spectroscopy )(Section )Molecular Spectroscopy : the interaction of electromagnetic radiation (light) with matter (organic compounds). Thisinteraction gives specific structural information. : Mass Spectrometry: molecular weight of the sample formulaThe mass spectrometer gives the mass to charge ratio (m/z), therefore the sample (analyte) must be an spectrometry is a gas phase technique- the sample mustbe vaporized.
2 Electron-impact ionizationSample Inlet10-7 - 10-8 torrR-Helectron beam 70 eV (6700 KJ/mol)e_R-H+massanalyzerm/zionization chamber(M+) u23 mass mcharge z==B2 r22VB= magnetic field strengthr = radius of the analyzer tubeV= voltage (accelerator plate)The Mass SpectrometerIonizationchamberIons of selectedmass/charge ratioare detectedIons of non-selectedmass/charge ratioare not detectedMagnetic Field, Bo4 Exact Masses of Common Natural IsotopesIsotope mass natural ( ) ( ) ( ) ( )Isotope mass natural abundance19F ( )79Br (98%) Ion (parent ion, M) = molecular mass of the analyte;sample minus an electronBase peak- largest (most abundant) peak in a mass spectra; arbitrarily assigned a relative abundance of 100%.
3 C6H6m/z = (M+)(100%)m/z=79 (M+1)(~ of M+)6 The radical cation (M+ ) is unstable and will fragment into smaller ionsRelative abundance (%)m/z=16 (M+)m/z=15m/z=14m/z=17 (M+1)CHHH+H+m/z = 15charge neutralnot detectedCH+HHcharge neutralnot detectedm/z = 14 CHHHH- e_CHHHH+m/z = 16+Relative abundance (%)m/zm/zm/z=15m/z=29m/z=43m/z=45 (M+1)m/z=44 (M)CHHCCHHHHHHCHHCCHHHHHH+CHHCCHHHHH+H+c harge neutral notdetectedm/z = 43m/z = 44 CHHCCHHHHHH- e_+CHHCHHH+CHHH+CHHCHHH+CHHH+m/z = 29m/z = 15charge neutralnot detectedcharge neutralnot detected- e_47m/zm/zClBrm/z=112(M+)m/z=113(M+ +1)m/z=114(M+ +2)m/z=115(M+ +3)m/z=7735Cl ( )79Br (98%)m/z=77m/z=156(M+)m/z=158(M+ +2)m/z=157(M+ +1)m/z=159(M+ +3)8 Mass spectra can be quite complicated and interpretation functional groups have characteristic fragmentationIt is difficult to assign an entire structure based only on the mass spectra.
4 However, the mass spectra gives the mass and formula of the sample which is very important obtain the formula, the molecular ion must be ionization techniquesMethods have been developed to get large molecules such as polymers and biological macromolecules (proteins, peptides, nucleic acids) into the vapor : Molecular Formula as a Clue to StructureNitrogen rule: In general, small organic molecules with anodd mass must have an odd number of nitrogens. Organic molecules with an even mass have zero or aneven number of nitrogens If the mass can be determined accurately enough, then the molecular formula can be determined (high-resolution mass spectrometry)Information can be obtained from the molecular formula:Degrees of unsaturation.
5 The number of rings and/or -bonds in a molecule (Index of Hydrogen Deficiency) 10 Degrees of unsaturationsaturated hydrocarbonCnH2n+2cycloalkane (1 ring)CnH2nalkene (1 -bond)CnH2nalkyne (2 -bonds)CnH2n-2 For each ring or -bond, -2H from the formula of the saturated alkaneHHHHHHHHHHHH C6H14- C6H12 H2 2 = 112 C6H14- C6H6 H8 8 = 412 HHHHHHH ydrogen DeficiencyDegrees of Unsaturation611 Correction for other elements:For Group VII elements (halogens): subtract 1H from the H-deficiency for each halogen, For Group VI elements (O and S): No correction is neededFor Group V elements (N and P): add 1H to the H-deficiency for each N or PC12H4O2Cl4 C10H14N2 : Principles of molecular Spectroscopy :Electromagnetic radiation = distance of one wave = frequency: waves per unit time (sec-1, Hz)c = speed of light ( x 108 m sec-1)h = Plank s constant ( x 10-34 J sec)Electromagnetic radiation has the properties of a particle (photon) and a (ground state)lighth organicmolecule(excited state)organicmolecule(ground state)+ h relaxation713h c Quantum: the energy of a photonE = h =E = c 10-1010-8!
6 -rays10-6x-rays10-5 UVVis10-4IR10-21microwavesradiowaves108 (cm)shorthighhighWavelength ( )Frequency ( )Energy (E)longlowlowE E 1 1 : Principles of molecular Spectroscopy :Quantized Energy Levelsmolecules have discrete energy levels (no continuum between levels)A molecule absorbs electromagnetic radiation when the energy of photon corresponds to the difference in energy between two states815UV-Vis: valance electron transitions - gives information about -bonds and conjugated systems Infrared: molecular vibrations (stretches, bends)- identify functional groupsRadiowaves: nuclear spin in a magnetic field (NMR)- gives a map of the H and C frameworkorganicmolecule(ground state)lighth organicmolecule(excited state)organicmolecule(ground state)+ h ultraviolet - visible (UV-Vis) Spectroscopy 200UV400800 nmVisRecall bonding of a -bond from Chapter -molecular orbitals of butadiene 1: 0 Nodes 3 bonding interactions 0 antibonding interactionsBONDING MO 2: 1 Nodes 2 bonding interactions 1 antibonding interactions BONDING MO 3: 2 Nodes 1 bonding interactions 2 antibonding interactions ANTIBONDING MO 4.
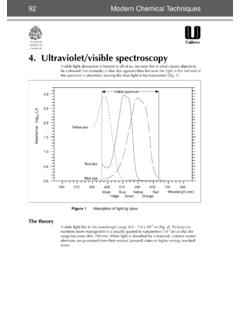
7 3 Nodes 0 bonding interactions 3 antibonding interactions ANTIBONDING MO 2 is the Highest Occupied Molecular Orbital (HOMO) 3 is the Lowest Unoccupied Molecular Orbital (LUMO)18UV-Vis light causes electrons in lower energy molecular orbitalsto be promoted to higher energy molecular LUMOC hromophore: light absorbing portion of a moleculeButadieneButadiene1019 Molecular orbitals of conjugated polyenesBondingAntibondingEnergyH2 CCH2180 nm217 nm258 nm290 nm20violet400 nmyellowblue450orangeblue-green500redyel low-green530red-violetyellow550violetora nge600blue-greenred700greenColor ofabsorbed light Colorobserved380 nm780 nm400 nm450 nm500 nm550 nm600 nm700 nmredorangeyellowgreenblueviolet-indigoM olecules with extended conjugation move toward the visible region1121 NNNNMgOCO2CH3 OOChlorophyll -carotenelycopeneOHOOHOHOOHOHOHOOHOHOH+a nthocyaninMany natural pigments have conjugated systems22 Chromophore: light absorbing portion of a molecule Beer s Law.
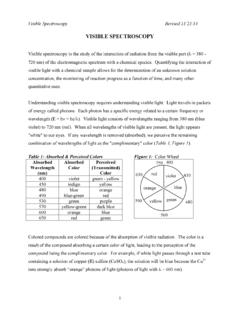
8 There is a linear relationship between absorbance and concentrationA = c lA = absorbancec = concentration (M, mol/L)l = sample path length (cm) = molar absorptivity (extinction coefficient) a proportionality constant for a specific absorbance of a : Introduction to Infrared SpectroscopyE 1 is expressed as (wavenumber), reciprocal cm (cm-1)_ =1 E _therefore_ (cm)VisNearIRFarIRInfrared (IR) x 10-4 x 10-3 cm16 m10-410-2!_4000!_600IR radiation causes changes in a molecular vibrations24 Symmetric stretchAntisymmetric stretchStretch: change in bond bendout-of-plane bendscissoringrockingwagging twistingBend: change in bond angle>>>>>>>>>>>>>>>>Animation of bond streches and bends: Stretch:Hooke s Law!
9 =_2 " c1mx mymx + myf12XY! = vibrational frequencyc = speed of lightmx = mass of Xmy = mass of Y_mx mymx + my= reduced mass ( )f = spring constant; type of bond between X and Y (single, double or triple)E f_Hooke s law simulation: of an Infrared Spectra:organic molecules contain many atoms. As a result, there are many stretching and bending modes- IR spectra have many absorption bandsFour distinct regions of an IR spectra 4000 cm-1600 cm-11500 cm-1fingerprintregiondoublebondregion200 0 cm-12500 cm-1triplebondregionX-Hsingle bondregionC-HO-HN-HC CC NC=CC=O1427 Fingerprint region (600 - 1500 cm-1)- low energy single bondstretching and bending modes.
10 The fingerprint region is unique for any given organic compound. However, there arefew diagnostic regions (1500 - 2000 cm-1)C=C 1620 - 1680 cm-1 C=O 1680 - 1790 cm-1 Triple-bond region: (2000 - 2500 cm-1)C C 2100 - 2200 cm-1 (weak, often not observed)C N 2240 - 2280 cm-1X-H Single-bond region (2500 - 4000 cm-1)O-H 3200 - 3600 cm-1 (broad)CO-OH 2500-3600 cm-1 (very broad)N-H 3350 - 3500 cm-1C-H 2800 - 3300 cm-1sp3 -C-H2850 - 2950 cm-1sp2 =C-H3000 - 3100 cm-1sp C-H3310 - 3320 cm-128 Alkenes=C-H3020 - 3100 cm-1medium - strongC=C1640 - 1680 cm-1 mediumAromatic=C-H3030 cm-1 strongC=C1450 - 1600 cm-1strongAlkynes C-H3300 cm-1strongC C2100-2260 cm-1weak - mediumAlcoholsC-O1050 - 1150 cm-1 strongO-H3400 - 3600 cm-1 strong and broadAminesC-N1030 - 1230 cm-1mediumN-H3300 - 3500 cm-1mediumCarbonylC=O1670 - 1780 cm-1strongCarboxylic acidsO-H2500 - 3500 cm-1strong and very broadNitrile C N 2240 - 2280 Characteristic Absorption FrequenciesTable , p.